Getting started
Many example workflows can be found when you first open Quasar or on the Orange website (https://orange.biolab.si/workflows/) including:
- Data and file table
- Hierarchical Clustering
- Principle Component Analysis
- Clustering
- Tile loader
Another good resource is the widget catalog. This information is also available in Quasar as built-in help, which you can access by selecting any widget and pressing F1.
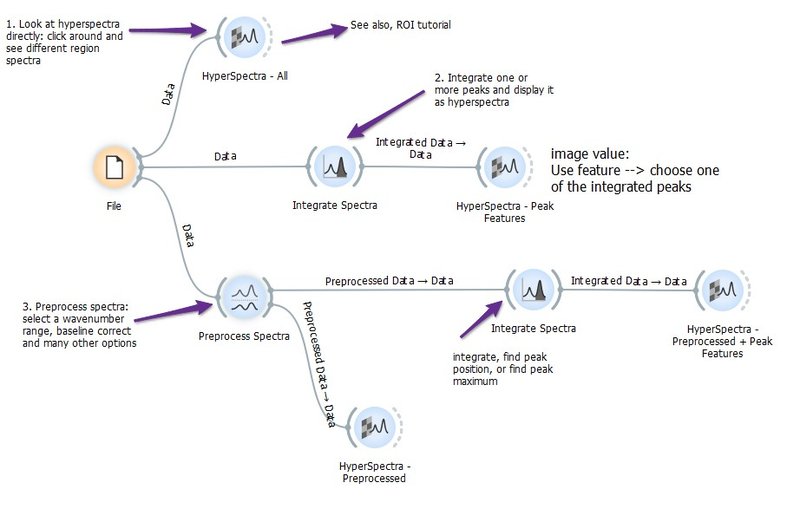
Exploring Hyperspectra
This example workflow shows some of the ways you can explore hyperspectral data.
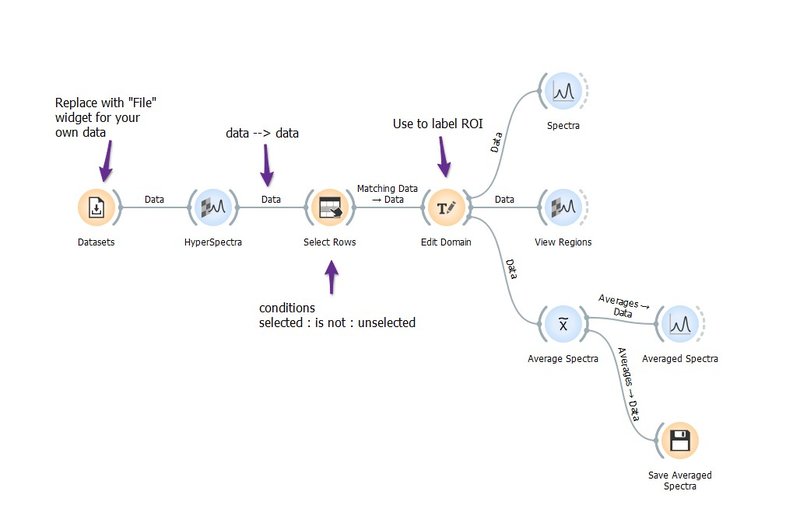
Selecting Regions of Interest
This example workflow demonstrates selecting multiple regions in hyperspectral maps and analyzing the spectra from those regions.
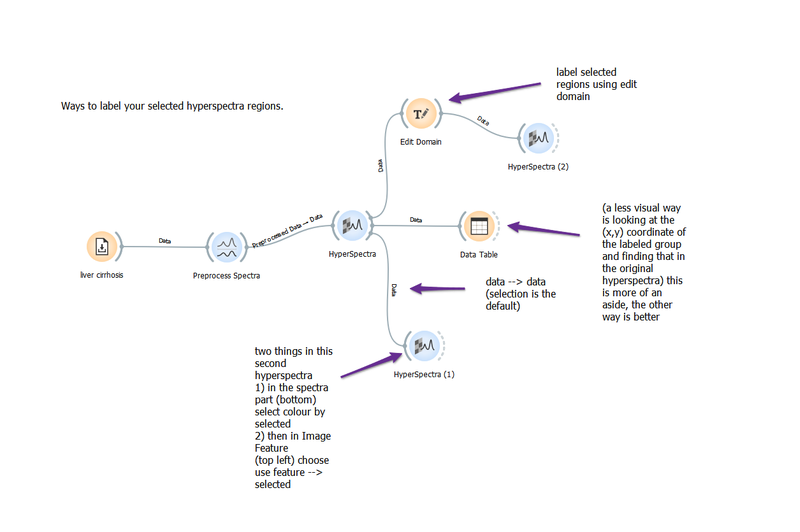
Labelling ROI in hyperspectra
Ways to label regions selected in a hyperspectra. This workflow can be used to label regions that you want to visualize in further analysis, for example PCA.
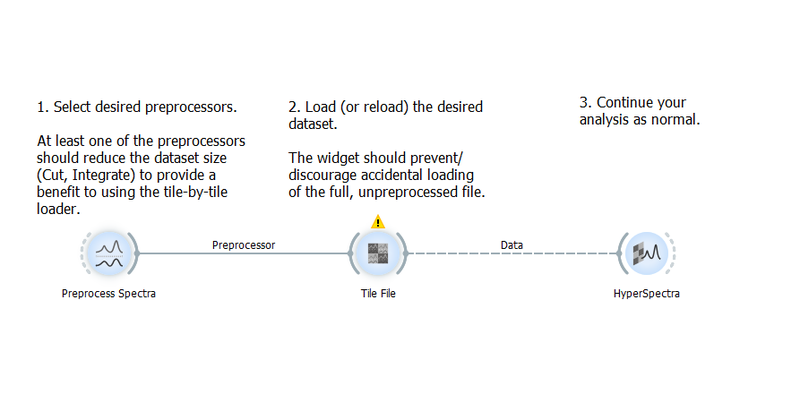
Large Data Sets
For large datasets, the tile loader is especially useful. The workflow demonstrates how to combine the tile-by-tile loader with preprocessors to load a large dataset directly into a reduced (and therefore smaller) form. It is useful for datasets where the file size is causing problems with memory usage and a preprocessor workflow has already been established.
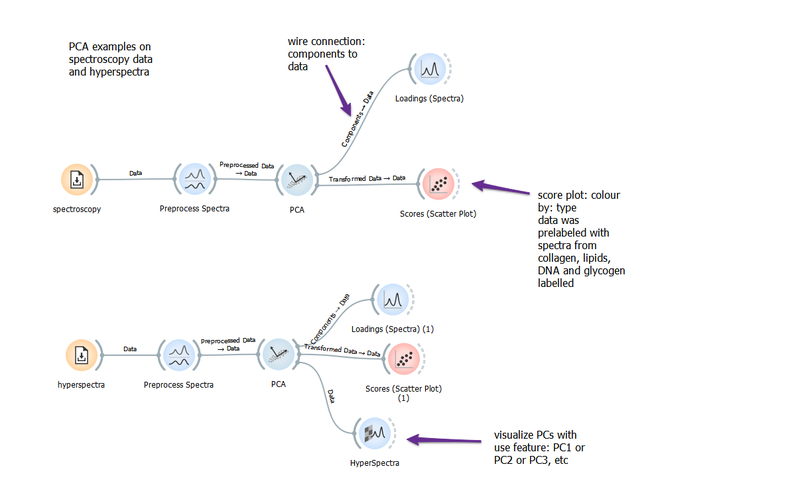
Basic PCA
PCA is a great way to reduce the dimensionality of hyperspectra data. This basic workflow shows how to make the wire connections in Quasar to visualize the scores, loadings and PCs as features in hyperspectra. Care must be taken when preprocessing data!